|
Protein Dynamics in Their Native Environment
In proteins, chemical or photochemical reactions can induce conformational changes, which alter the chemical and physical properties of these molecules. In the physiological environment of a cell these altered properties may represent a `signal', like in the case of receptors, or may trigger the function, like in the cases of ion channels or enzymes. The altered properties can also represent a malfunction or a disease like in the case of the conversion of prion proteins into their scrapie isoform (TSE diseases).
Our main goal within the field of Theoretical Molecular Biophysics is to develop concepts
and (computational) methods for improved descriptions of such processes and for the analysis
of corresponding experimental observations (see our molecular dynamics package EGO MMII). As to explore the viability of these methods we study a carefully selected set of applications in close cooperation with experimental groups.
|  |
|
Virtual Single Molecule Spectroscopy in Condensed Phase
|
 |
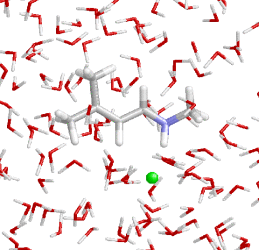
The complex structure and dynamics of proteins strongly alters the properties of embedded molecules both in the ground and excited electronic states. In particular optical spectra and photochemical reactions of chromophores like retinal in bacteriorhodopsin are steered by the structured electrostatic field in the binding pocket. Similarly the ground state force fields and, thus, the vibrational spectra are strongly affected by such fields.
We are engaged in further development of hybrid methods which combine quantum mechanical (QM) descriptions of molecules with molecular mechanics (MM) simulations of their environment. In a series of applications these QM/MM methods (see our molecular dynamics package EGO MMII) are employed to study selected functional processes in proteins (see also Prof. Nonella). Also here close cooperations with spectroscopists are vital.
|
Data based Modelling and Neuroinformatics
We study relations between learning in neural tissue and algorithms for statististical data analysis. Here the motivation is our belief that nature has invented not only seemingly complicated organisms but also elegant and simple solutions for complicated problems of information processing and pattern recognition. Thus, corresponding algorithms must not be invented but rather may be found by analysis of brain function. As main software tools MULTIVAR and NEURONET so far emerged from this effort.
|
|
|